Comparative Analysis of Five Commercial RT-PCR Diagnostic Assay for Detection of Covid-19
Abstract
SARS-CoV-2 real-time reverse-transcription PCR (rRT-PCR) is the most effective testing system available to combat COVID-19, given the absence of any specific treatment or vaccine. Moreover, numerous SARS-CoV-2 rRT-PCR kits have been approved under emergency-use-authorization (EUA) worldwide. In this article, we present a comparison of important performance features among five commercial RT-PCR assays.
A total of consecutive nasopharyngeal (NPS) samples and oropharyngeal (OP) swabs were collected from 50 COVID-19 patients to analyze sensitivity and specificity. The results showed variations in sensitivity among all the RT-PCR kits examined. The Pishtaz teb assays demonstrated the highest positive percent agreement (PPA) of 95.2% (40/42), followed by Covitech (90.5% - 38/42), DaAn Gene (83.3% - 35/42), Sansure (66.66% - 28/42), and Power check of SARS- CoV-2 panel (64.3% - 27/42).
Conversely, all five molecular assays demonstrated a negative percent agreement (NPA) of 100% (8/8). These findings provide a technical baseline for assessing the performance of five distinct commercial PCR assays for detecting SARS-CoV-2. They could prove practical and useful for laboratories seeking to purchase any of these assays for further clinical validation.
Highlights
·Compared five COVID-19 RT-PCR kits approved and available by Iran Ministry of Health.
·Pishtaz teb's kit identified the highest number of positive clinical samples.
Author Contributions
Academic Editor: John Akighir, Federal University Wukari, Taraba State, Nigeria
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2023 Sepideh Hasanzadeh, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an infectious disease that emerged in the Middle East 1. It is a concerning public health issue due to its worldwide spread and unexpectedly high mortality rate 2. SARS-CoV-2 is constantly scattered around the world, and as of October 21st, 2020, over 41 million cases have been confirmed globally, causing over 1,130,496 deaths 3. At the time of writing this paper, the Centers for Disease Control and Prevention (CDC) reported more than 539,670 confirmed COVID-19 cases and 31,034 deaths in Iran 3, 4.
Recently, this epidemic has placed a heavy burden on society, the global economy, and health care systems all around the world. Several measures are being taken to control its prevalence and incidence 5. Many of these measures depend on the accurate diagnosis of infected people. Reverse transcription-polymerase chain reaction (RT-PCR) based on detecting and quantifying a fluorescent signal is the most sensitive and specific method; therefore, it is the most preferred test 6. Although different SARS-CoV-2 RT-PCR kits are commercially accessible, an independent assessment and comparison of these products are urgently needed to guide the accuracy of tests in a diagnostic market filled with new tests 7. Although the RT-qPCR assay was attended as the gold-standard method for identification of respiratory viruses such as SARS-CoV and Middle Eastern respiratory syndrome coronavirus (MERS-CoV), current RT-PCR assays targeting SARS-CoV-2 have some flaws and limitations 8. Two main challenges are encountered with the initial use of these assays. Firstly, due to the high genetic similarity between SARS-CoV-2 and SARS-CoV, there is cross-reactivity of the primers and probes used. Secondly, these assays may lack sufficient sensitivity to confirm infections in suspicious patients during the early stages after admission. Notably, there have been reported cases where CT scan results showed signs of infection, but the RT-PCR assay yielded negative results upon the first presentation 9.
Coronaviruses are enveloped positive-sense RNA viruses that express their replication and transcription complex, such as open-reading frame (ORF1a and ORF1b), RNA-dependent RNA polymerase (RdRp). Moreover, they express the coronavirus structural proteins, including the envelope (E), nucleocapsid (N), and spike (S) proteins, by the transcription of subgenomic messenger RNAs during replication cycle stages of the far outnumber (anti) genomic RNAs. The ORF1ab/RdRp, E, N, and S genes were consequently developed and targeted regions in the SARS-CoV-2 genome 5. To identify coronavirus disease of 2019 (COVID-19), the World Health Organization (WHO) suggests the E gene as the first-line screening, the RdRp gene as the confirmatory assay, and the N gene as an additional confirmatory assay 10. For example, the European Virus Archive GLOBAL (EVAg) primer-probe set targets the E and RdRp regions, the DAAN kit targets the ORF1ab and N coding regions, and the BGI kit targets the ORF1ab region 11, 12, 13.
In the early stages of the outbreak, both international and national agencies rushed to begin mass production of test reagents and issued an Emergency Use Authorization (EUA) for the CDC COVID-19 RT-PCR method. Despite these efforts, laboratories still face several problems, such as a shortage of reagents, lack of tools access, inability to perform high-complexity tests, and an increase in staffing needs. These issues leave a gap in health care providers' ability to quickly diagnose and manage patients. The urge to find a sensitive, available, and rapid diagnostic test for the detection of COVID-19 is obvious. Here, we have compared five COVID-19 RT-PCR kits that have been approved and are available by the Ministry of Health in Iran to evaluate their sensitivity and specificity.
Material and methods
The study was conducted at Imam Reza Hospital in Mashhad, Iran between March and October 2020. We selected various RT-PCR commercial assays based on specific criteria: i) The assay can be performed on standard real-time PCR thermocyclers available worldwide, ii) The assay must be available in the market or the test can be made available by the manufacturer in the pre-release version, iii) The assay must be registered by the National Medical Device Directorate of IR. IRAN (IMED). Table 1 presents the selected assays and their details.
Table 1. Overview of kits for RT-PCR-based detection of SARS-COV-2 included in the study| Manufacturer | Country | Storage condition | Target gene(s) | Instruments Channels | Ct value |
| Sansure | China | -20 ±5℃ | ORF1ab, N | FAM, ROX | ≤ 40 |
| DaAn Gene | China | -20±5℃ | ORF1ab, N | VIC, FAM | ≤ 40 |
| Power check | Korea | -25–15℃ | RdRP, E | FAM, JOE | ≤ 37 |
| Pishtaz teb | Iran | -20±5℃ | RdRp, N | FAM, HEX | ≤ 40 |
Our main focus was to evaluate the rate of false negatives (FN) reports and the sensitivity of each assay using 50 COVID-19 patient samples. To this end, both nasopharyngeal (NP) and oropharyngeal (OP) swabs were collected from 50 hospitalized patients at Imam Reza Hospital, who exhibited suspected COVID-19 symptoms. We stored the swabs in tubes containing 3 ml of Universal Transport Media.
To assess the efficiency and limit of detection (LOD) of each assay, we initially conducted a duplicate 10-fold dilution series of specified viral RNA. This RNA was isolated from SARS-CoV-2 viral particles obtained from the COVID-19 National Reference Lab of Pasteur Institute of Iran, using the RNJia Virus Kit. We calculated the slope through linear regression using GraphPad Prism.
For the sample size calculation, we employed PASS 11.0.8 software, considering a paired-sample sensitivity power analysis with a power of 80%, type I error of 5%, sensitivity of 100% versus 65% (for two types of diagnostic kits), and discordant proportion of 0.35. The calculated required sample size, with a 10% attrition rate, was 50.
Subsequently, we prepared a panel of 50 clinical samples and extracted RNA from these samples using the RNJia Virus Kit. All PCRs were conducted on a Rotorgene II (Qiagen) following the manufacturer’s instructions. Each sample was run in duplicate and repeated three times over three consecutive days. The results were analyzed, interpreted as positive (Ct<40) or negative (Ct>40), and recorded along with the Ct value of each target gene.
Statistical analysis, including the comparison of Ct values, was performed using the SPSS software program, version 15. A P-value of <0.05 was considered statistically significant.
Result
The PCR Efficiency of Commercial Assays
We initially assessed the PCR efficiency for each target gene assay by conducting a duplicate 10-fold dilution series of SARS-CoV-2 specified viral RNA (Figure 1). All RT-PCR kits exhibited PCR efficiencies greater than 80%. Notably, the Pishtaz teb and DaAn gene assays demonstrated the highest efficiency, at approximately 92%. Meanwhile, the Covitech and Powercheck assays exhibited efficiencies of 87% and 85.39%, respectively. The RdRp gene assay of Sansure displayed the lowest efficiency, measured at 80.44% (Table 2).
Table 2. Sensitivity and specificity of the PCR kits.| Company | Sensitivity | Specificity | Accuracy |
|---|---|---|---|
| Pishtaz teb | 95.2% | 100% | 96% |
| Da An Gene | 83.3% | 100% | 86% |
| Sansure | 66.7% | 100% | 72% |
| Power check | 64.3% | 100% | 70% |
| Covitech | 90.5% | 100% | 92% |
Figure 1.PCR Efficiency Assessment for Four Commercially Available RT-PCR Kits for SARS-CoV-2 RNA Detection: To determine the PCR efficiency (E) of each target gene, we employed a duplicate 10-fold dilution series of SARS-CoV-2 viral RNA. Linear regression analysis was performed in GraphPad Prism to obtain the slope and R2 values. The percentage efficiency was then calculated from the slope using the formula E = 100 * (-1 + 10^(-1/slope)).
The clinical sensitivity of assays on clinical samples
After testing 50 clinical specimens, the Pishtaz teb assay demonstrated the highest Positive Percent Agreement (PPA) at 95.2% (40/42), followed by Covitech with 90.5% (38/42). The DaAn Gene, Sansure, and Power check SARS-CoV-2 panel showed PPAs of 83.3% (35/42), 66.7% (28/42), and 64.3% (27/42), respectively.
All five molecular assays exhibited a Negative Percent Agreement (NPA) of 100% (8/8). As Pishtaz teb detected the most positive samples (40/42), we compared all kits to Pishtaz teb. The results from Power check and Pishtaz teb showed that 53.1% of samples were detected by both assays, 14 samples were detected only by Pishtaz teb, and only one sample was detected by Power check. The p-value between these two assays was 0.001.
Similarly, the results of Sansure and Pishtaz teb demonstrated that only 57% of the samples were identified by both assays, with 13 tests detected only by Pishtaz teb and one sample detected by Sansure. The p-value between these two assays was 0.002. For Pishtaz teb and DaAn Gene, 60.7% of the results complemented each other and were close. The p-value between these two assays was 0.016. The Covitech results were closest to Pishtaz teb, missing only four positive samples that were identified by Pishtaz teb. The p-value was 0.5.
Based on the difference in detection rates for each gene, Pishtaz teb identified 11 positive samples in both channels, while 29 samples were identified only in the yellow channel (N gene). Samples were considered positive if repeated. Compared to Pishtaz teb, the Covitech diagnostic kit identified 12 positive samples in both channels, one more than Pishtaz teb, while 6 samples were identified in the yellow channel by Pishtaz teb but not by Covitech. The DaAn Gene diagnostic kit detected seven specimens in the green channel (N gene) that were only detected in the yellow channel by Pishtaz teb.
In contrast, Pishtaz teb identified seven specimens in the yellow channel that DaAn Gene could not detect. The Sansure kit detected four samples in the green channel (ORF1ab gene) that were identified only in the yellow channel by Pishtaz teb. Sansure also detected one sample in the yellow channel (N gene) that Pishtaz teb could not identify. However, Pishtaz teb identified eleven samples that Sansure could not detect. The PowerCheck demonstrated a good level of identification in the green channel (RdRp gene), identifying sixteen samples that Pishtaz teb found only in the yellow channel. Additionally, PowerCheck identified one sample in the yellow channel that Pishtaz teb did not detect (Table 3).
Table 3. Clinical performance comparison of four molecular assays for the detection of SARS-CoV-2| Company | No. Of Results | No. Detection rate Of Each Gene | ||||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | ND 1 | N | RDRP | E | Orf1ab | S | |
| Pishtaz teb | 40 | 8 | 2 | 40 | 11 | - | - | - |
| Da An Gene | 35 | 8 | 7 | 35 | - | - | 32 | - |
| Sansure | 28 | 8 | 14 | 28 | - | - | 15 | - |
| Power check | 27 | 8 | 15 | - | 16 | 27 | - | - |
| Covitech | 38 | 8 | 4 | - | - | 38 | - | 12 |
Conversely, eight samples were identified by Pishtaz Kit that PowerCheck did not identify. The difference in detection rates was significant between Pishtaz teb and the other kits (P-value<0.05), except for the yellow channel's results of the Sansure, which did not show a significant P-value. When CT values were considered in both channels, Pishtaz teb exhibited the lowest mean CT (Figure 2) with a significant difference from the mean CT of other assays (P < 0.001). The remaining four kits had statistically higher CT values compared to the Pishtaz teb assay.
Figure 2.Different RT-PCR kits showed variations in Ct values.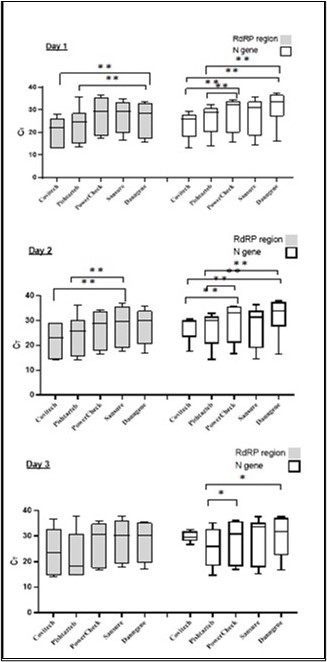
Discussion
Here we have provided a comparison of five commercially available RT-PCR kits for the detection of SARS-CoV-2 in clinical samples (n = 50). The Pishtaz teb’s kit and Covitech diagnosed 40, 38 positive results out of 50 samples, respectively, which is the highest number of positive clinical samples. Dangen kits were able to positively identify 35 out of 50 samples. Sansure and PowerCheck exhibited 14,15 negative results, which are the most; therefore, needs a further survey. Particularly, we performed our analysis with a small number of clinical specimens; thus, we advise that diagnostic laboratories conduct additional clinical examinations upon the application of novel RT-PCR kits. One of the advantages of all these diagnostic kits based on multiple target genes is that the results can be interpreted as those of a combination of target genes, which complements both sensitivity and specificity.
A major challenge encountered in standard real-time PCR analysis is false negative reports, caused by inhibitors or inefficient PCR conditions 14, 15. Internal controls are used to address reliability, by adding extra primer-probe sets to target other endogenous DNA sequences or exogenous targets 16, 17.
Conclusion
As a conclusion, we confirmed that all of the commercially available RT-PCR kits included in this study can be used for the diagnosis of patients with symptoms of COVID-19. When performing virus diagnostics tests in populations that low viral loads expected to display, such as health-care workers with mild or no symptoms or patients during the early stages of the infection, it might be advisable to use those kit with the best performance regarding the identification of clinical samples, i.e., RT-PCR kits from Pishtaz teb,Covitech and Dangene Which showed high identification rate among other kits.
Funding
This research has not received any external funding
Declaration of Interest
The authors declare no conflicts of interest.
Abbreviations
MERS-CoV: Middle Eastern Respiratory Syndrome Coronavirus
N: Nucleocapsid NP: Nasopharyngeal NPA: Negative Percent Agreement ORF: Open-Reading Frame OP: Oropharyngeal RT-PCR: Reverse transcription-polymerase chain reaction RdRp: rRNA-dependent RNA polymerase SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2 S: Spike WHO: World Health Organization.References
- 1.Fan W, Su Z, Bin Y, Yan-Mei C, Wen W et al.A new coronavirus associated with human respiratory disease in China. , Nature 579(7798), 265-9.
- 2.Raoofi A, Takian A, Sari A A, Olyaeemanesh A, Haghighi H et al.COVID-19 pandemic and comparative health policy learning in Iran. , Archives of Iranian Medicine 23(4), 220-34.
- 4.CfD Control, Prevention. (2020) National diabetes statistics report. Centers for Disease Control and Prevention, US Department of Health , Atlanta, GA: .
- 5.Corman V M, Landt O, Kaiser M, Molenkamp R, Meijer A et al.Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. , Eurosurveillance 25(3), 2000045.
- 6.Reusken C B, Broberg E K, Haagmans B, Meijer A, Corman V M et al. (2020) Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories. in 30 EU/EEA countries , Eurosurveillance 25(6), 2000082.
- 8.Oliveira B A, LCd Oliveira, Sabino E C, Okay T S.SARS-CoV-2 and the COVID-19 disease: a mini review on diagnostic methods. Revista do Instituto de Medicina Tropical de Sao Paulo. 2020-62.
- 9.Xie X, Zhong Z, Zhao W, Zheng C, Wang F et al. (2020) Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 200343.
- 10.Corman V, Bleicker T, Brünink S, Drosten C, Zambon M. (2020) Organization WH. Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR. Geneva: World Health Organization.
- 12.DAAN. (2019) Instructions for Use of Detection Kit for Novel Coronavirus (2019-nCoV) RNA (Fluorescent PCR). , Guangzhou, China:
- 14.Raymaekers M, Smets R, Maes B, Cartuyvels R.Checklist for optimization and validation of real‐time PCR assays. , Journal of clinical laboratory analysis 23(3), 145-51.
- 15.Hur K-H, Park K, Lim Y, Jeong Y S, Sung H et al.Evaluation of four commercial kits for SARS-CoV-2 real-time reverse-transcription polymerase chain reaction approved by emergency-use-authorization in Korea. Frontiers in Medicine. 2020, 521.
